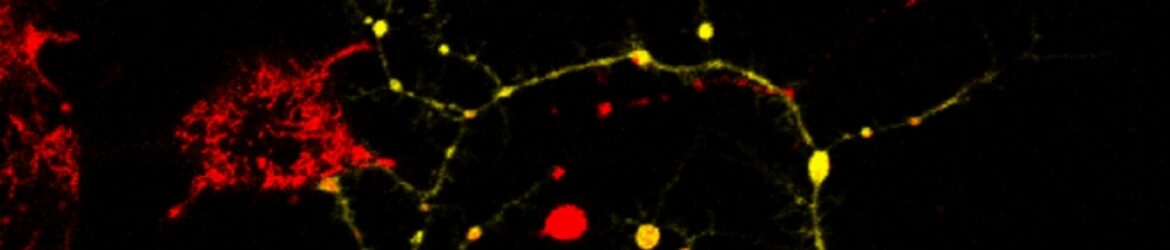
mRNA localization to mitochondria

Extensive research in our lab had identified molecular players that mediate the interaction of mRNA with yeast mitochondria. We found that the outer membrane protein Tom20 interacts with proteins’ nascent chains, thereby bringing translated mRNAs to mitochondria proximity, Furthermore, we found that the protein OM14 interacts with the ribosome-associated NAC complex, thereby further stabilize mRNA association with mitochondria.
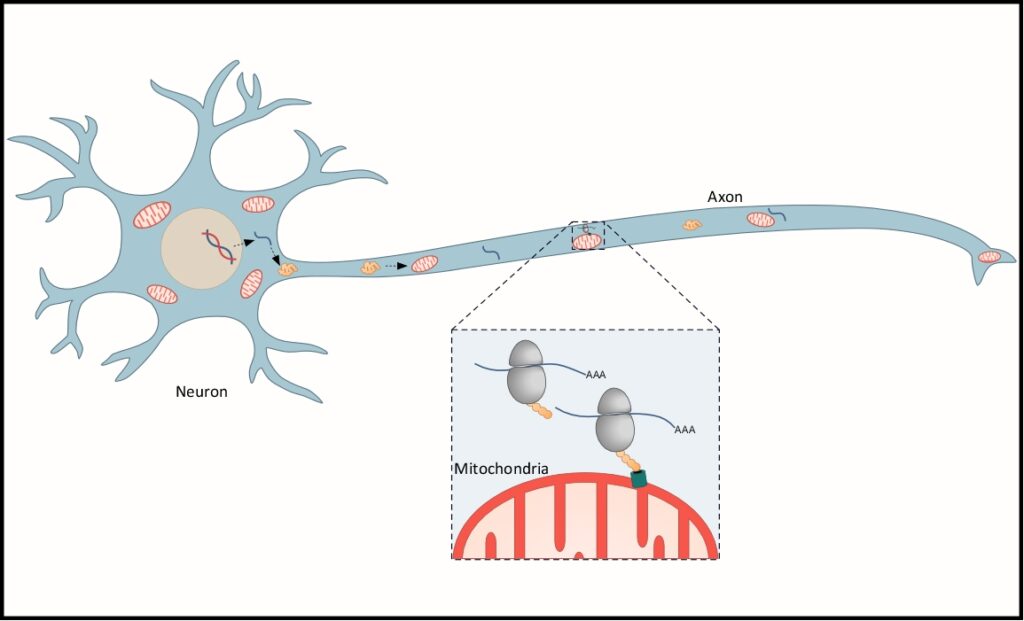
We now extend these finding to neurons, which heavily depend on mitochondrial activity. Defects in mitochondrial activity are implicated in many neurological disorders and diseases. Of significant challenge is the maintenance of mitochondrial activity at distant neuronal sites (e.g. synapses). It is not known yet how the many different proteins that function in mitochondria approach these distant sites. We explore whether mitochondria are associated with an active translation machinery and whether this plays a role in refueling distant mitochondria with new proteins.
RNA binding proteins in translation regulation
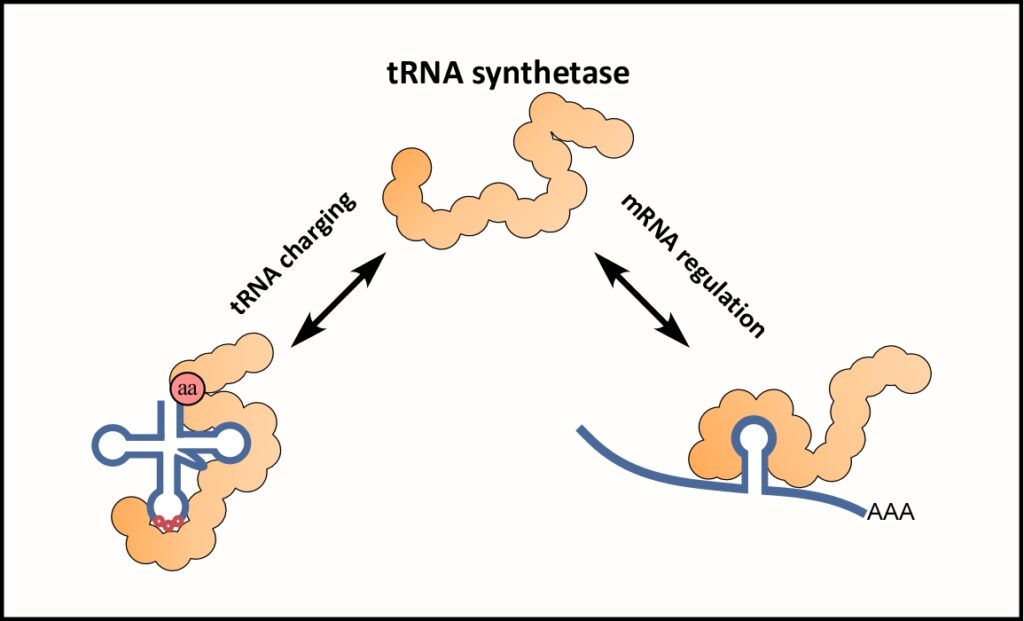
RNA binding proteins are central regulators of post-transcriptional processes. We study mRNA binding proteins that are involved in translation regulation and mRNA localization, using yeast as a model organism. We revealed unexpected modes of association between Pumillio members and mRNAs, as well as new and surprising mRNA binding proteins. Among these new binders, we currently investigate the possible roles of aminoacyl tRNA synthetases (aaRSs). These well-studied tRNA-binders were found to bind many types of mRNAs, through a mechanism that mimics their binding mode to tRNA. For example, we found that HisRS; the aaRS which charge tRNAHis with histidine, binds its own mRNA to regulate its translation according to tRNAHis levels and charging demands. Further research is aimed at identifying the role of RNA modifications in binding and translation regulation, and possible implications in diseases.